
Saulius Klimasauskas
Vilnius University
Engineering metabolic cascades for selective chemical tracking of epigenetic signaling in live cells
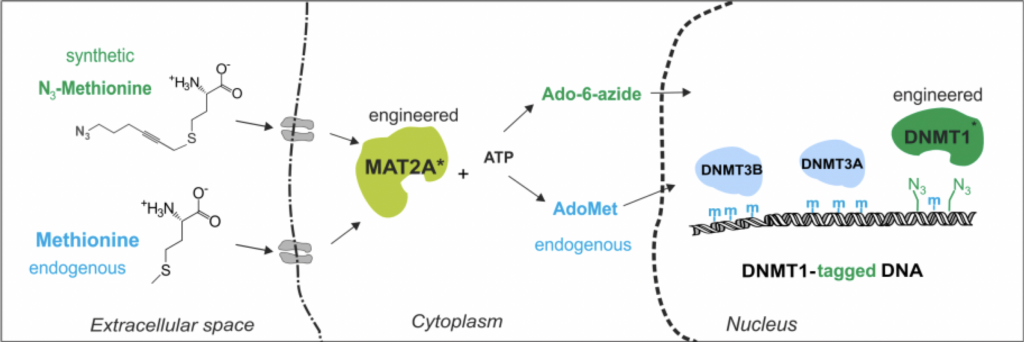
The ubiquitous biological methyl donor for transmethylation reactions, S-adenosyl-L-methionine (AdoMet), is produced from L-methionine (Met) and ATP by methionine adenosyltransferases (MAT). Methylation of cytosine to 5-methylcytosine in DNA is a fundamental epigenetic mechanism involved in mammalian development and disease. DNA methylation is brought about by collective action of three independently expressed and regulated AdoMet-dependent proteins (Dnmt1, Dnmt3A and Dnmt3B). However, the catalytic interactions and temporal interplay of these epigenetic “writers” in maintaining cell identity and cell fate commitment are still poorly understood. To achieve catalytic tracking of each individual Dnmt enzyme, we turned to structure-guided engineering of the mouse Dnmt1 and Dnmt3A MTases for the transfer of bioorthogonal 6-carbon linear moieties containing a functional azide group onto DNA from a synthetic cofactor analog, Ado-6-azide [1]. We then installed the corresponding codons in the Dnmt1 or Dnmt3A alleles using CRISPR-Cas9 genome editing of mouse ESCs and used electroporation for pulse-internalization of the Ado-6-azide cofactor into the engineered cells to achieve selective catalysis-dependent azide-tagging of Dnmt-specific targets in vivo [2]. Most recently, we established in situ production of Ado-6-azide cofactor by similarly engineering the genomic allele of a key MAT enzyme, MAT2A, in mouse ESCs to enable efficient conversion of a cell-penetrant synthetic extended L-methionine analog, N3-Met. This permitted chemically controlled biorthogonal labeling of genomic Dnmt targets in doubly-engineered mouse cells under mild stress-free conditions. The deposited azide tags can be exploited as “click” handles for precise mapping of the tagged methylation sites in the genomic sequence [2] or spatial visualization in fixed nuclei. Altogether, we demonstrate the first engineered enzymatic cascade that produces high-resolution chemical “tracks” of the Dnmt1 or Dnmt3A catalysis in live mammalian cells. Our newly developed general approach permits temporal monitoring of the individual epigenetic writers during the cell cycle or cell state transitions, offering unprecedented inroads into epigenetic one-carbon signalling pathways in a wide range of eukaryotic model systems.
 __________
__________
[1] G. Vilkaitis, V. Masevičius, E. Kriukienė, S. Klimašauskas. Acc. Chem. Res., 2023, 56: 3188-3197.
[2] V. Stankevičius, P. Gibas, B. Masiulionytė, L. Gasiulė, V. Masevičius, S. Klimašauskas, G. Vilkaitis. Mol. Cell, 2022, 82: 1053-1065.
_________
View Abstract as PDF:
_________
Tue. April 16 | 09:50 – 10:10 hrs – Engineering metabolic cascades for selective chemical tracking of epigenetic signaling in live cells
